
Home page of STOP lab
Genome Regulation Centre

Welcome to the STOP lab
Genome Regulation – focusing on Gene Ends
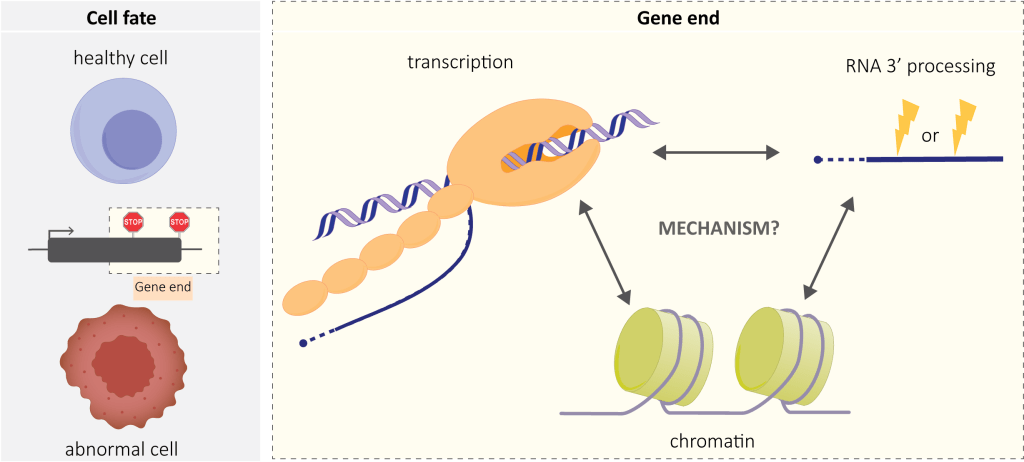
Our lab’s aim is to integrate different levels of gene expression regulation – chromatin, transcription and RNA processing – focusing on gene ends.
We want to understand the basic mechanisms of when and how transcription stops (terminates), but also how perturbed termination contributes to disease. Premature termination, called also transcription attenuation, is particularly relevant in the pathological context. Premature termination was known to be an important regulatory mechanism in bacteria and yeast, but overlooked in humans and animals in general. We and others have recently shown that this phenomenon is a very abundant genomic event in metazoa. It has also medical implications, particularly in cancer.
Our main approaches are based on NGS (genomics and nascent transcriptomics), combined with molecular biology, biochemistry and single molecule RNA fluorescent in situ hybridization. The lab’s experimental workhorse are mammalian cell culture models of cancer and neuronal differentiation. We use both experimental and computational approaches.
We are always interested in new team members – if you are enthusiastic, motivated and potentially willing to apply for external funding, please get in touch with Kinga.
Our team
PI

Kinga Kamieniarz-Gdula, PhD, DSc
Group leader
kinga.kamieniarz-gdula [at] amu.edu.pl
Kinga finished her undergraduate studies at Adam Mickiewicz University in Poznań. She then did her PhD as well as a short postdoc in the field of Chromatin and Epigenetics with Rob Schneider at the Max-Planck-Institute in Freiburg, Germany. After that she moved to the University of Oxford to join Nick Proudfoot’s lab, supported by a Marie Skłodowska-Curie Fellowship. In Oxford, Kinga started studying the links between transcription termination and RNA processing, and became fascinated by gene ends.
After 13 years abroad, Kinga returned to Poznań to set up her own research group in October 2019. Her team is generously supported by NAWA Polish Returns, NCN SONATA BIS, EMBO Installation Grant and ERC Starting and Proof of Concept Grants.
Besides science Kinga enjoys reading non-fiction, playing the piano, zumba and hiking with her family.
Agata Stępień, PhD, eng.

Agata received her Engineer and MSc degree in biotechnology from the University of Agriculture in Krakow. She then moved to Poznan for her PhD, and worked in the group of Prof. Jarmolowski and Prof. Szweykowska-Kulinska at Adam Mickiewicz University. Her PhD research was focused on the characterization of the crosstalk between the spliceosome and the microprocessor in plants by molecular biology, microscopic and biochemical techniques. After graduating in 2017, Agata got her first Postdoc position in K. Dorota Raczynska’s group to study the role of U7 snRNA in human cells. In 2019, Agata joined STOP lab to investigate transcription termination changes during cancer progression.
Martyna Plens-Gałąska, PhD

Martyna received her MSc degree in Molecular Biology from Adam Mickiewicz University in Poznań where she continued with PhD research in the group of prof. Hans Bluyssen. She focused on potential strategies to inhibit STAT proteins activity in inflammation during onset and progression of atherosclerosis. Currently she is writing up her PhD thesis and making sure experiments in STOP lab can run smoothly.
Martyna loves cats, dogs and plants and aims to convert our hallway to an exotic greenhouse. In her free time you’ll find her in the bushes watching birds.
Joanna Kwiatkowska, PhD

Asia earned her MSc and PhD degrees in Molecular Biology from the Adam Mickiewicz University. In the Laboratory of RNA Biochemistry, headed by Prof. Mikolaj Olejniczak, she used biochemical approaches to characterize the mechanism of bacterial RNA molecules interaction with chaperone proteins. She joined STOP lab in June 2022 and is interested in biochemical aspects of alternative polyadenylation. She likes non-fiction literature, jogging and sewing.
Deepshika Pulimamidi, MSc

Deepshika obtained her Master’s degree in Bioinformatics from Pondicherry University in India. Following her postgraduation she gained experience in two different labs in India where her focus was to understand the novel crosstalk between histone post-translational modifications (specifically methylation) and RNA polymerase II mediated gene expression. In her research, she applied NGS techniques such as RNA-Seq and ChIP-Seq to understand how epigenetics and transcription regulatory pathways are interconnected to regulate gene expression patterns, as they often tend to overdrive in disorders like cancer. In April 2023, Deepshika joined STOP lab to study the cross-talk of RNA cleavage and transcription termination. Besides research, she is passionate about dancing, travelling and cooking.
Apoorva Shrivastava, MSc

Apoorva completed his integrated Bachelor and Master of Technology dual degree in Biotechnology from KIIT University, India. He worked at a genome sequencing company before moving to Birmingham, UK. In order to learn in-depth about molecular biology, he pursued MSc in Molecular Biotechnology from University of Birmingham where he also worked under Dr. Matthias Soller for a short duration, studying m6A methyltransferase complex. He joined STOP lab in December 2024 under the ERC grant and is currently working on Understanding the crosstalk between 3’ end processing and transcription termination from genetic perspective. If not in lab, you would find him playing badminton or critically analysing movies. He is also fond of photography and runs a dedicated page for photography on instagram.
Magda Kopczyńska, MSc

Magda graduated from the Poznan University of Medical Sciences with a Master’s degree in Medical Biotechnology. During her undergraduate studies she worked on the role of long non-coding RNA molecules in cancer and their potential utility as biomarkers at the Laboratory of Cancer Genetics (Greater Poland Cancer Center). In October 2020 she started her PhD studies in our team and investigates how the chromatin environment influences transcription termination, and vice versa. In her spare time Magda enjoys popular science writing. Together with Martyna they form the green-fingered fraction in the lab, taking loving care of the plants on our floor.
Upasana Saha, PhD

Upasana received her MSc in Biotechnology from Visva-Bharati in India. She continued her PhD in India at Jadavpur University under the supervision of Prof. Biswadip Das. Her research involved understanding mRNA quality control pathways in Saccharomyces cerevisieae, focusing mainly on the nucleus. In 2019, she moved to the lab of Prof. Torben Heick Jensen in Aarhus University, Denmark for her first postdoc position. There she studied the role of the nuclear poly(A) binding protein Nab2p in mRNA homeostasis. She joined STOP lab in October 2021 to investigate the molecular mechanism of premature transcription termination. Upasana loves cooking, shopping and exploring new places.
Ewa Stein, PhD

Ewa graduated from Adam Mickiewicz University in Poznań with a MSc in Biotechnology. She also started her PhD there in the Laboratory of RNA Biochemistry in the group of prof. Mikołaj Olejniczak. Her research concerned the recognition of bacterial small RNAs by proteins using biochemical methods. In the STOP lab Ewa assists the project led by Asia.
Ewa likes reading and spending time outdoors.
Klaudia Ostatek, MSc

Klaudia is a molecular biologist and biotechnology researcher. She completed her bachelor’s and master’s studies at TU Dresden, Germany—first in German, then in English—and conducted her master’s thesis at Stockholm University in Sweden. In October 2024, she began her PhD studies in our team. Her research focuses on transcription termination, particularly on enhancing the expression of the limiting factor PCF11. She is also involved in smFISH (Single Molecule Fluorescence In Situ Hybridization) experiments to visualize transcription and RNA localization. Outside the lab, she enjoys learning foreign languages, photography, film editing, traveling, and caring for her ever-growing collection of plants. She hopes to incorporate lymphatic tissue research into her scientific career in the future.
Selected publications
1) SETD2 methyltransferase activity aids gene definition by promoting correct transcription initiation and termination.
Nakayama C, Kopczyńska M [joint-first author], Stępień A, Ito S, Imami K, Gdula MR, Nojima T, Kamieniarz-Gdula K.
(BioRxiv preprint) 2025 https://doi.org/10.1101/2025.07.14.664659
2) A beginners guide to Sf9 and Sf21 insect cell line culture and troubleshooting.
Kwiatkowska J, Stein E, Romanenko A, Plens-Gałąska M, Podsiadła-Białoskórska M, Szołajska E, Kühn U, Kamieniarz-Gdula K.
Sci Rep. 2025 https://doi.org/10.1038/s41598-025-99812-0
3) Defining gene ends: RNA polymerase II CTD threonine 4 phosphorylation marks transcription termination regions genome-wide.
Kopczyńska M, Saha U, Romanenko A, Nojima T, Gdula MR, Kamieniarz-Gdula K.
Nucleic Acids Research. 2025 https://doi.org/10.1093/nar/gkae1240
4) CLP1-dependent premature transcription termination opposes neurodegeneration.
Gdula MR, Kopczyńska M, Saha U, Kamieniarz-Gdula K.
Neuron. 2022 https://doi.org/10.1016/j.neuron.2022.03.012
5) Transcriptional Control by Premature Termination: A Forgotten Mechanism.
Kamieniarz-Gdula K, Proudfoot NJ.
Trends Genet. 2019 https://doi.org/10.1016/j.tig.2019.05.005
6) Selective Roles of Vertebrate PCF11 in Premature and Full-Length Transcript Termination.
Kamieniarz-Gdula K, Gdula MR, Panser K, Nojima T, Monks J, Wiśniewski JR, Riepsaame J, Brockdorff N, Pauli A, Proudfoot NJ.
Mol Cell. 2019 https://doi.org/10.1016/j.molcel.2019.01.027
7) WNK1 kinase and the termination factor PCF11 connect nuclear mRNA export with transcription.
Volanakis A, Kamieniarz-Gdula K [joint-first & co-corresponding author], Schlackow M, Proudfoot NJ.
Genes Dev. 2017 https://doi.org/10.1101/gad.303677.117
8) BRCA1 recruitment to transcriptional pause sites is required for R-loop-driven DNA damage repair.
Hatchi E, Skourti-Stathaki K, Ventz S, Pinello L, Yen A, Kamieniarz-Gdula K, Dimitrov S, Pathania S, McKinney KM, Eaton ML, Kellis M, Hill SJ, Parmigiani G, Proudfoot NJ, Livingston DM.
Mol Cell. 2015 https://doi.org/10.1016/j.molcel.2015.01.011
9) R-loops induce repressive chromatin marks over mammalian gene terminators.
Skourti-Stathaki K, Kamieniarz-Gdula K, Proudfoot NJ.
Nature. 2014 https://doi.org/10.1038/nature13787
10) The genomic landscape of the somatic linker histone subtypes H1.1 to H1.5 in human cells.
Izzo A, Kamieniarz-Gdula K [joint-first author], Ramírez F, Noureen N, Kind J, Manke T, van Steensel B, Schneider R.
Cell Rep. 2013 https://doi.org/10.1016/j.celrep.2013.05.003
11) Regulation of transcription through acetylation of H3K122 on the lateral surface of the histone octamer.
Tropberger P, Pott S, Keller C, Kamieniarz-Gdula K, Caron M, Richter F, Li G, Mittler G, Liu ET, Bühler M, Margueron R, Schneider R.
Cell. 2013 https://doi.org/10.1016/j.cell.2013.01.032
12) A dual role of linker histone H1.4 Lys 34 acetylation in transcriptional activation.
Kamieniarz K, Izzo A, Dundr M, Tropberger P, Ozretic L, Kirfel J, Scheer E, Tropel P, Wisniewski JR, Tora L, Viville S, Buettner R, Schneider R.
Genes Dev. 2012 https://doi.org/10.1101/gad.182014.111
More publications in PubMed.
Collaborations
Prof. Elmar Wahle, MLU, Halle, Germany
Dr Takayuki Nojima, Kyushu University, Japan
Dr Stephan Hamperl, HMGU, Munich, Germany
Dr Shazia Irshad, University of Oxford, UK
Prof. Hiroshi Kimura, Tokyo Institute of Technology, Japan
Recent Tweets
YouTube: popular science
Funding

Find us
Lab location

We are affiliated with the Institite of Molecular Biology and Biotechnology (IBMiB), Faculty of Biology, Adam Mickiewicz University (AMU) in Poznań. Our laboratory is located in the building of the Center for Advanced Technology, at the Morasko Campus which hosts all Science Faculties of AMU.
Poznań

Poznań is the capital of the Wielkopolska (Greater Poland) region. It is a vibrant university city, with an urban area population of ~1 million, located only 3 hours away from Warsaw and Berlin. At the same time, it is a paradise for nature lovers, as it is surrounded by beautiful forests & lakes.
Contact

Address:
Center for Advanced Technology
Uniwersytetu Poznańskiego 10
61-614 Poznań, POLAND
Email: kinga.kamieniarz-gdula [at] amu.edu.pl
Lab members: name.surname@amu.edu.pl